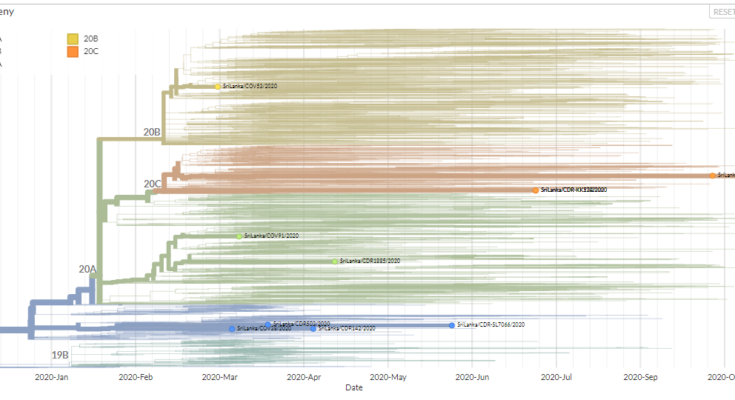
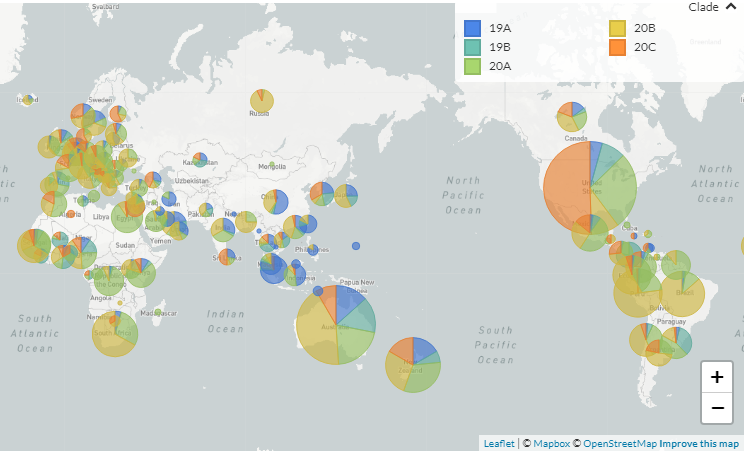
Next Generation Sequencing (NGS) analysis of SARS CoV 2 viral samples by the Center for Dengue Research of Sri Jayewardenepura University has shown the presence of four clades out of five reported clades- 19A, 19B, 20A, 20B and 20C of Corona virus in Sri Lanka. Last week the laboratory announced the identification of the viral strain 20C as the viral strain behind the ongoing COVID 19 outbreak. The Clade 20C was first identified in Sri Lanka on July 27th according to a sequencing data available on nexstrain.org website where they keep and analyze genome sequencing data of SARS CoV 2 viral strains from all around the globe.
Clade 20C “The American Clade” ?
A clade is a strain of virus having same source of origin or characteristics (mutations). According to nexstrain.org researchers so far have identified five different clades of the SARS CoV2 virus. Their origin and unique mutations are as follows,
| Clade | Primary Countries | Mutations | Max Frequency |
| 19A | Asia: China/Thailand | Root clade | 65-47% Globally in Jan |
| 19B | Asia: China | C8782T T28144C | 28-33% Globally in Jan |
| 20A | N America/Europe/Asia: USA, Belgium, India | C14408T A23403G | 41-46% Globally Apr-May |
| 20B | Europe: UK, Belgium, Sweden | G28881A G28882A G28883C | 19-20% Globally Mar-Apr |
| 20C | N America: USA | C1059T G25563T | 19-21% Globally Apr |
You can view the current clades on the Global SARS-CoV-2 Nextstrain tree here.
As you can see in the above table, the first two clades were named as 19A and 19B which correspond to the split marked by mutations C8782T and T28144C. These clades were both prevalent in Asia during the first months of the outbreak. The next clade that was named is 20A corresponding to the clade that dominated large European outbreak in early 2020. It is distinguished from its parent 19A by the mutations C3037T, C14408T and A23403G.

After this, we’ve seen two further clades appear: 20B, another European clade separated clearly by three consecutive mutations: G28881A, G28882A, and G28883C and 20C, a largely North American clade, distinguished by mutations C1059T and G25563T.
Out of these five clades only four have been detected in Sri Lanka, as there have been no reports of Clade 19B in Sri Lanka. According to the recent sequencing data, the viral clade behind the ongoing outbreak belongs to the 20C clade which had been originated in USA. When the outbreak began in September many speculations arose about from where this virus came and how? Many thought the virus came to Sri Lanka from India, as it’s our neighboring country and an ongoing huge outbreak of the virus was happening in India at that time period. According to researchers the sequencing data or identifying the exact clade alone is not enough to decide the source of a viral infection.
According to the Pangolin Lineage tool which is another widely used mapping system to describe and identify different mutations of SARS nCoV 2 virus genome, this new virus strain is found to be from B.1.42 lineage which had been described as the Sweden and Denmark lineage.
Center for Dengue Research Undertaking the COVID 19 Sequencing as a National Duty!
Established in 2012 the Center for Dengue Research (CDR) is one of the main research laboratories that undertook research activities related to the Dengue viral infections in Sri Lanka. But with the emergence of COVID 19 pandemic, the research unit began helping the national effort of controlling the pandemic in Sri Lanka by utilizing their human and physical resources for RT-PCR testing and viral genome sequencing. Currently, the CDR is the only laboratory in Sri Lanka that carries out genome sequencing for the COVID 19 virus samples using the state of art new NGS technology they possess. As of today, the unit has shared results of 25 COVID 19 virus sequencing data with nextrain.org where they have analyzed the data and mapped in the Global SARS-CoV-2 Nextstrain tree. The CDR functions under the Department of Immunology and Molecular medicine of Univeristy of Sri Jayawardenapura.
Sequencing the source of a pandemic; Why it’s important?
Speaking to the Sri Lankan Scientist magazine, Prof. Neelika Malavige, the director of the CDR, and also a member of the team sequencing this virus, explained the importance of the sequencing data of disease-causing viruses.

University of Sri Jayawardenapura
“One of the main characteristics of a disease causing new virus is that they may mutate very rapidly, and these mutations can change the way the virus behaves inside our bodies. So, it’s really important to keep track of these mutations so that we can get an idea about the changes of the virulence and infectivity like parameters of a virus. This is why we started in March itself. However, it is now clear that the SARS-CoV2 is a very stable virus, with just around 2 mutations/month” . Basically, it is important to know there is only one SARS-CoV2 virus causing COVID-19 and small changes in the nucleotide sequence would assign these viruses to different lineages.”
“And also having an idea about the exact strain of the virus that is spreading in a community is also very much important for many epidemiological processes. For an example, if a virus has many different strains circulating, we can suspect a high level of community spread. And some countries use sequencing in contact tracing as well.”
Prof. Neelika further talked about the importance of recent sequencing outcomes as well,
“Due to the influx from individuals from different countries in the world, we saw circulation of different strains in March. In the CMC cluster in April, we saw several strains responsible for this. However, the Kandakadu cluster was due to one strain only. The current circulating virus strain is the of the B.1.42 lineage, which is similar to those seen in Europe. However, this does not mean the virus came from Europe. Just that this virus strain was first identified from Europe and was the predominant virus strain in some European countries. It has the mutation (D614G) associated with high transmissibility due to high viral loads, which is the dominant strain in the world right now. It was inevitable that we would get this strain sooner or later. Unfortunately, due to this mutation, it is associated with rapid spread and therefore, it is imperative that public take more precautions. Actually, many countries recommend a distance of 2 m instead of 1 m to reduce the spread. Therefore, its important for proper hand hygiene, wearing masks and avoiding unnecessary movement .”

The team of scientists that carried out the sequencing work are Dr. Chandima Jeewandara, Dr. Deshni Jayathilaka, Dr. Dinuka Ariyaratne, Mr. Laksiri Gomes and Mr. Diyanath Ranasinghe led by Prof. Neelika Malavige. Dr. Ananda Wijewickram and Dr. Malika Karunaratne from NIID, provided the initial samples from the Brandix cluster for sequencing.